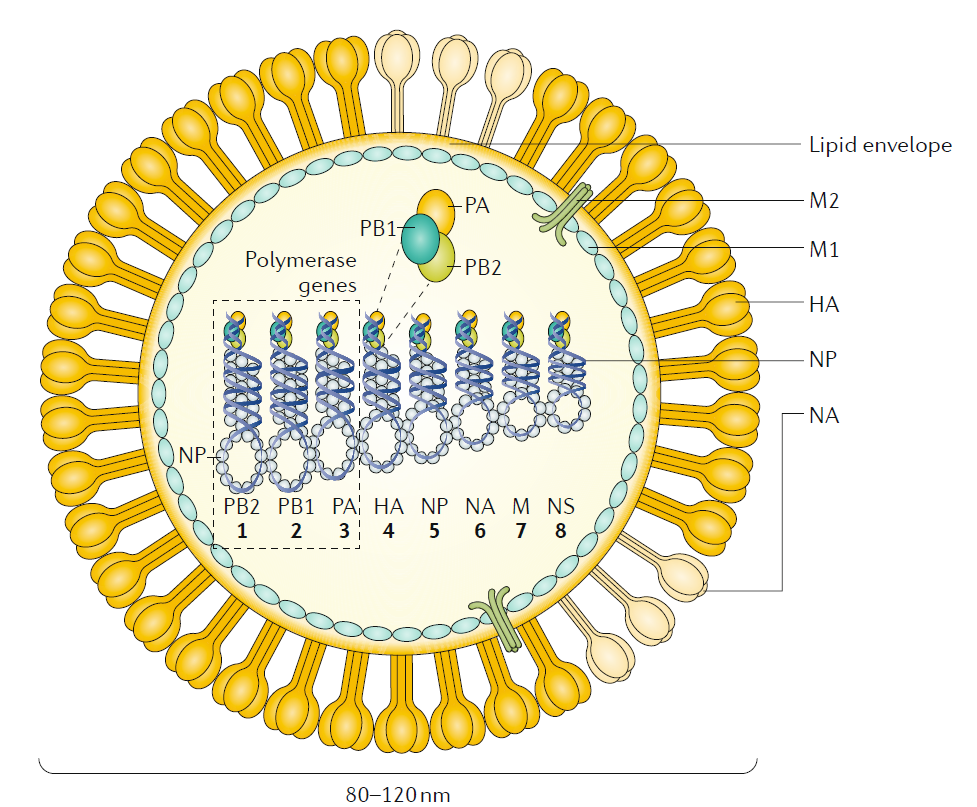
 |
Figure 1 Influenza A and influenza B. The figure represents an influenza A virus particle or virion. Both influenza A and influenza B viruses are enveloped negative- sense RNA viruses with genomes comprising eight single- stranded RNA segments located inside the virus particle. Although antigenically different, the viral proteins encoded by the viral genome of influenza A and influenza B viruses have similar functions: the three largest RNA
segments encode the three subunits of the viral RNA- dependent RNA polymerases (PB1, PB2 and PA) that are responsible for RNA synthesis and replication in infected cells; two RNA segments encode the viral glycoproteins haemagglutinin (HA , which has a ‘stalk’ domain and a ‘head’ domain), which mediates binding to sialic acid- containing receptors and viral entry , and neuraminidase (NA), which is responsible for releasing viruses bound to non- functional receptors and helping viral spread. The RNA genome is bound by the viral nucleoprotein (NP), which is encoded by RNA segment 5. RNA
segments 6 and 8 encode more than one protein, namely , the matrix protein (M1) and membrane protein (M2) — BM2 in the case of influenza B — and the nonstructural protein NS1 (not shown) and nuclear export protein (NEP). The M1 protein is thought to provide a scaffold that helps the structure of the virion and that, together with NEP, regulates the trafficking of the viral RNA segments in the cell; the M2 protein is a proton ion channel that is required for viral entry and exit and that, together with the HA and NA glycoproteins, is located on the surface of the virus anchored in a lipid membrane derived from the infected cell. Finally , the NS1 protein is a virulence factor that inhibits host antiviral responses in infected cells. The influenza viruses can also express additional
accessory viral proteins in infected cells, such as PB1–F2 and PA- x (influenza A), that participate in preventing host innate antiviral responses together with the NS1 protein or NB (influenza B), the function of which is unknown. NS1, NEP, PB1–F2 and PA- x are not present in the virus particle or are present in only very small amounts. NB is a unique influenza B virus surface protein anchored in the lipid membrane of the virus particles.
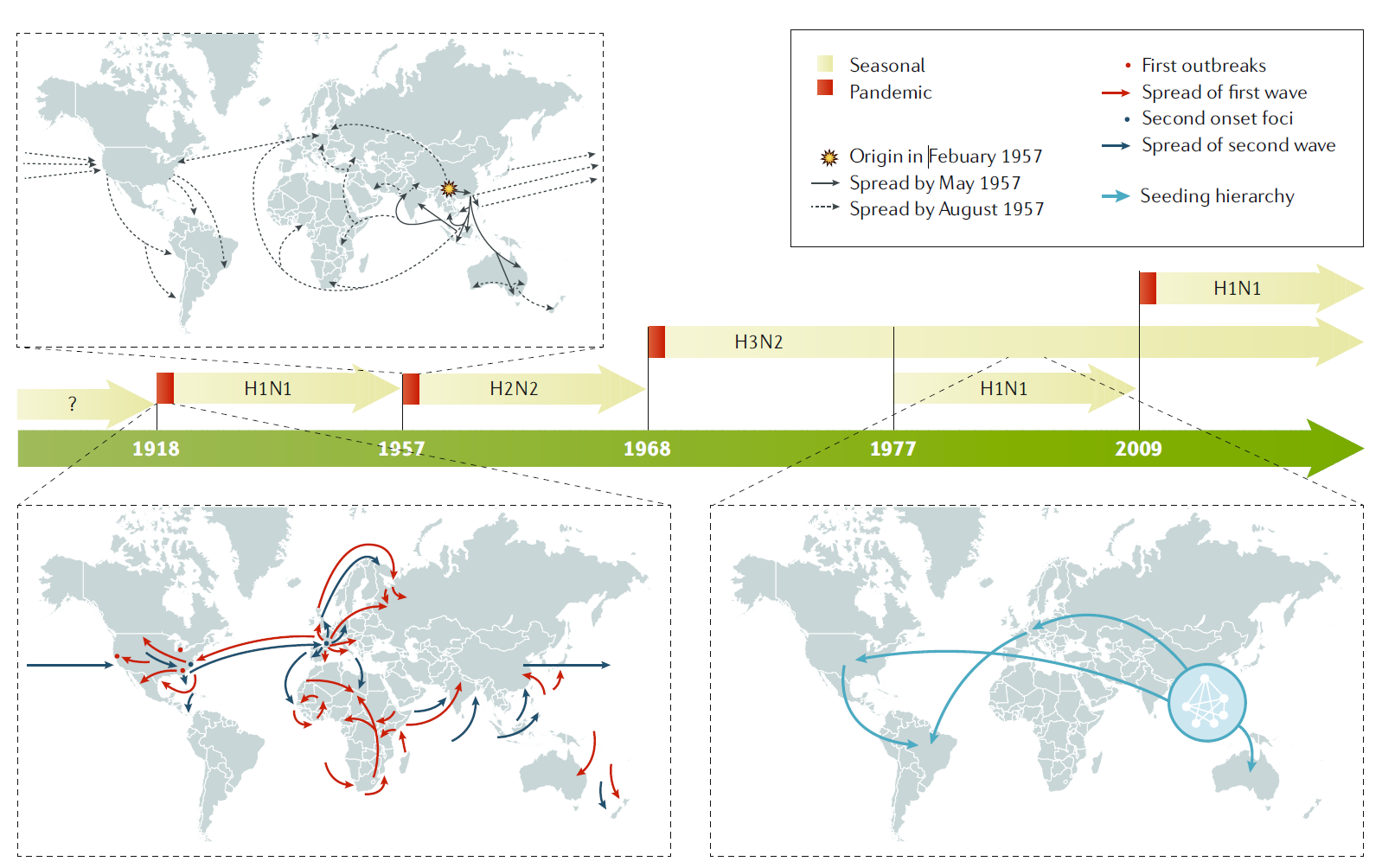
Figure 2 Influenza pandemics. In the past 100 years, four pandemics of human influenza have occurred, with the 1918
pandemic caused by an influenza A H1N1 virus being the most devastating, as it was associated with >40 million deaths.
Influenza A H2N2, H3N2 and H1N1 viruses caused the 1957 , 1968 and 2009 pandemics, respectively. In 1977 , influenza A H1N1 restarted circulation in humans without causing a pandemic, as the strain was similar to that which preceded the 1957 influenza A H2N2 pandemic. By contrast, the 2009 pandemic influenza A H1N1 virus was antigenically very different to the previous seasonal influenza A H1N1 viruses and replaced them as the circulating influenza A H1N1 strain. Examples of the spreading of human influenza A viruses in the world are shown for pandemic 1918 and 1957 viruses and for seasonal H3N2 viruses18. For pandemic virus outbreaks, the arrows indicate the first and second waves of transmission. For seasonal influenza A H3N2 spread, the arrows indicate the seeding hierarchy of seasonal influenza A (H3N2) viruses over a 5-year period, starting from a network of major cities in east and southeast Asia; the hierarchy within the city network is unknown. Seasonal influenza B viruses (not shown) are co- circulating in humans with influenza A viruses. |
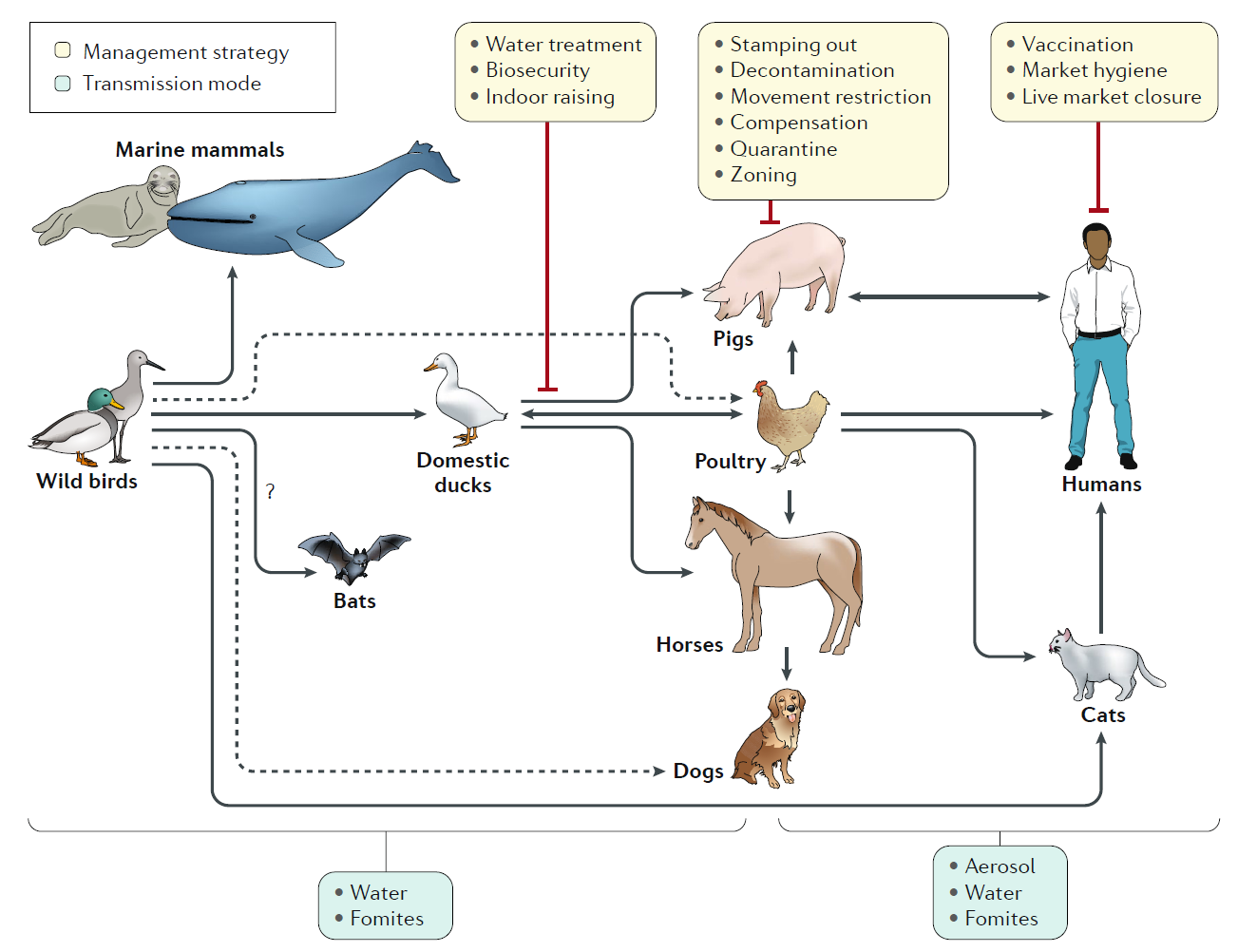
Fig. 3 | Emergence of influenza A virus from aquatic wild bird reservoirs. Influenza A viruses have been found in multiple species all seemingly derived from viral ancestors in wild birds, with the possible exception of bat influenza- like virus, which is of still uncertain origin. Influenza viruses from wild birds can spill over through water or fomites to marine mammals and to domestic free- range ducks. Transmissions to other avian species (for example, poultry) from domestic ducks or directly from wild birds can also occur from contaminated water. Transmission from ducks to other species occurs through ‘backyard’ farming, whereby the animals are raised together, and in live poultry and/or animal markets. Transmission from backyard to commercial farms can occur via lack of biosecurity and via spread through live markets. Humans can be infected with poultry and swine influenza viruses through aerosols, fomites or contaminated water.
However, in most instances these infections do not result in subsequent human- to-human transmission. Human- to-human transmission of seasonal or pandemic human viruses can be mediated by respiratory droplets, aerosols or self- inoculation after touching of fomites. Additional virus adaptations would be required for sustainable human- to-human transmission of animal influenza viruses. Other domestic animals known to be susceptible to influenza virus infections are dogs and cats. Dashed lines represent transmission that bypasses a domestic duck intermediate. |
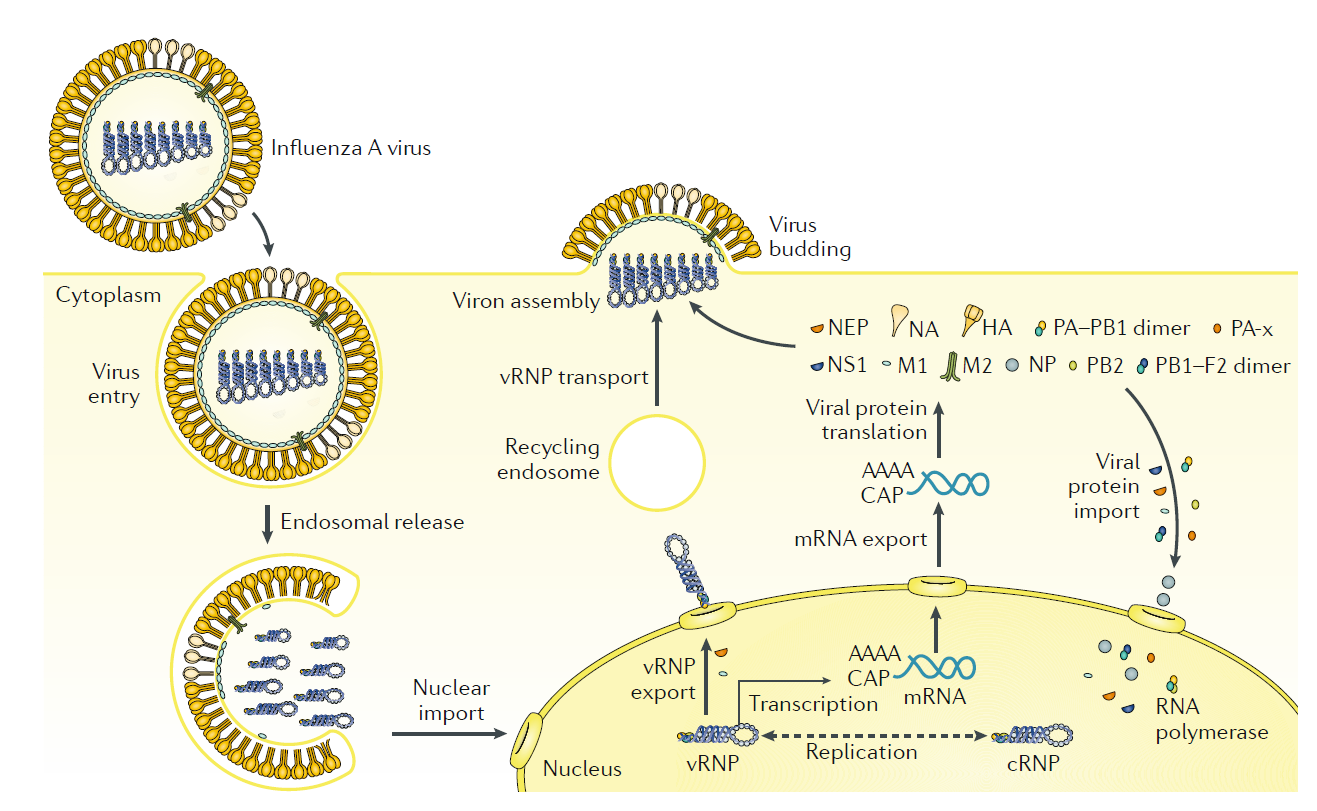 |
Fig. 4 | Influenza virus life cycle. Influenza virus enters the cell by endosomal uptake and release, and its negative- sense genetic material in the form of viral ribonucleoproteins (vRNPs) is imported to the nucleus for transcription of mRNA and replication through a positive-sense complementary ribonucleoprotein (cRNP) intermediate. Viral mRNA is translated into viral proteins in the cytoplasm, and these are assembled into new virions together with the newly synthesized vRNPs. PB1–F2 is shown here as a dimer, but can also be multimeric. HA , haemagglutinin; M1, matrix protein; M2, membrane protein; NA , neuraminidase; NEP, nuclear export protein; NP, nucleoprotein; NS1, nonstructural protein; PB1, PB2 and PA , viral RNA polymerases.
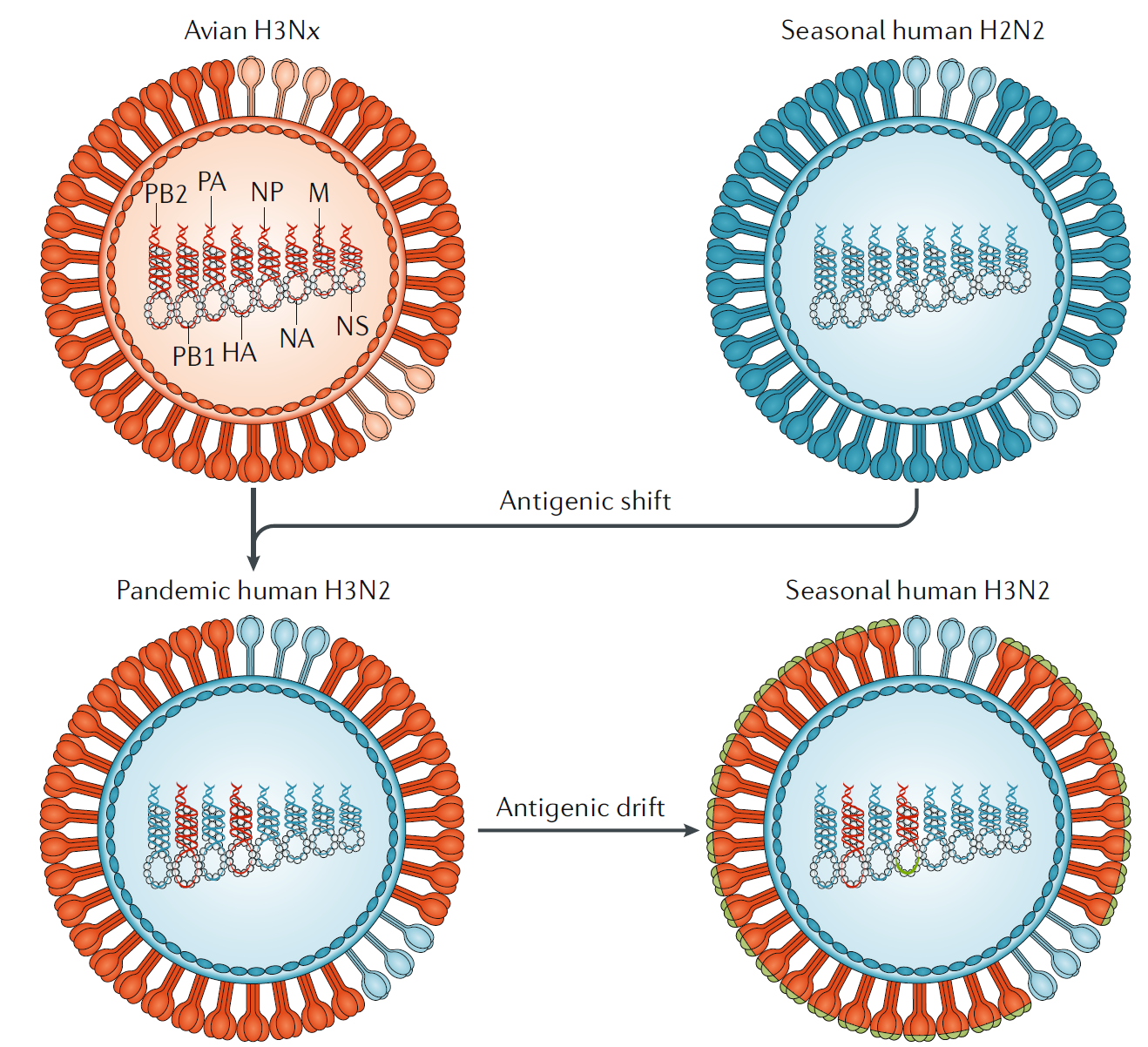
Fig. 5 | Influenza antigenic shift and antigenic drift. In 1968, co- infections between an avian influenza A H3Nx (where x = 1–9) virus and the seasonal human influenza A H2N2 viruses resulted in the exchange of viral segments (reassortment) and the selection of the pandemic human influenza A H3N2 virus, with the RNA polymerase PB1 and haemagglutinin (HA) RNA segments derived from the avian virus and the rest of the segments derived from the human virus. The lack of pre- existing immunity to the antigenically novel H3 HA in humans facilitated human transmission. Similar reassortment processes have taken place during other influenza A virus pandemics. Once H3N2 became established in humans, the virus began to drift, as is the case with all other human seasonal influenza viruses. During drift, small antigenic changes in the HA protein generated by mutation are selected to increase immune evasion, although not as dramatically as during shift. M, RNA encoding M1, matrix protein, and M2, membrane protein; NA , RNA encoding neuraminidase; NP, RNA encoding nucleoprotein; NS1, RNA encoding nonstructural protein; PB2 and PA , RNA encoding RNA polymerases.
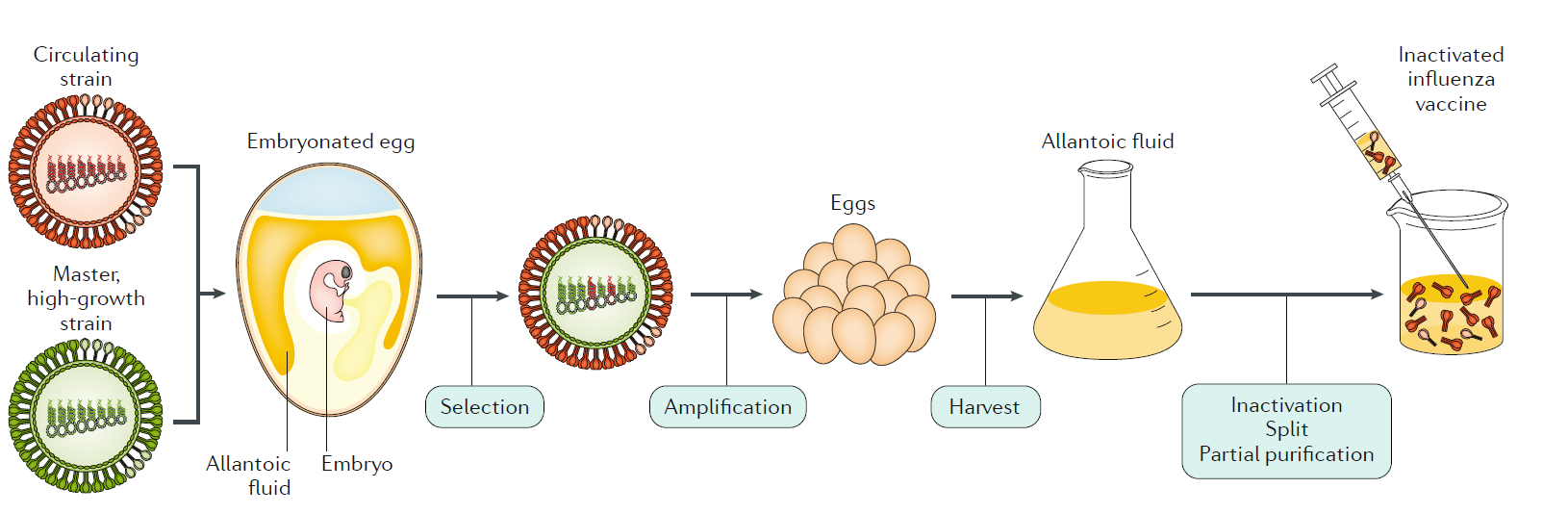
Fig. 7 | Inactivated influenza A virus vaccine manufacture. An antigenically representative circulating strain is reassorted with the high- growth influenza A/Puerto Rico/8/1934 (PR8) virus strain by co- infection in eggs, and a vaccine virus is selected with the high- growth properties of PR8 (conferred by its RNA segments encoding internal viral proteins) and the haemagglutinin (HA) and neuraminidase (NA) derived from the circulating strain. The vaccine virus is amplified in eggs, and the allantoic fluid from the infected eggs containing high titres of the virus is harvested; the virus is inactivated, treated for splitting of individual viral components and subjected to partial purification to enrich for the HA (and NA) viral components in the final injectable vaccine.
Influenza, Nature Reviews, 2018
The epidemic influenza virus is commonly referred to as the influenza virus. It is divided into types A, B, and C, and the recently discovered influenza virus is categorized as type D. The influenza virus can infect and cause disease in various animals, such as humans, poultry, pigs, horses, and bats. It is the pathogen for human influenza, avian influenza, swine influenza, equine influenza, and other zoonotic diseases.
The epidemic influenza virus (influenza virus) is a representative species of the Orthomyxoviridae family. It includes human and animal influenza viruses. Human influenza viruses are divided into types A, B, and C and are the pathogens for epidemic influenza. The antigenicity of the Type A influenza virus often undergoes variations and has caused several global pandemics. For instance, during the 1918-1919 pandemic, at least 20 to 40 million people worldwide died from influenza. Type B influenza virus also has significant pathogenicity to humans, but there has not been a global pandemic caused by it. Type C influenza virus only causes mild or unnoticeable upper respiratory infections in humans and rarely becomes epidemic. The Type A influenza virus was successfully isolated in 1933, Type B in 1940, and Type C not until 1949.
Influenza Classification Based on the hosts the influenza virus infects, it can be categorized as human influenza virus, swine influenza virus, equine influenza virus, and avian influenza virus. The human influenza viruses can be further divided into three classes based on their nucleoprotein antigenicity:
- Influenza A virus
- Influenza B virus
- Influenza C virus The influenza viruses that infect birds, pigs, and other animals have nucleoprotein antigenicity identical to that of human Type A influenza, but since the classification of Types A, B, and C only pertains to human influenza, avian influenza and the like are not typically referred to as Type A. Based on nucleoprotein antigenicity, influenza viruses are also divided into different subtypes according to hemagglutinin (HA) and neuraminidase (NA) antigenicity.
Morphology The influenza virus is spherical, with newly isolated strains often being filamentous. Its diameter is between 80 and 120 nanometers, while the length of filamentous influenza can reach up to 4000 nanometers. The structure of the influenza virus can be divided into three parts: the envelope, matrix protein, and core.
Core The virus core contains the genetic material storing virus information and the enzymes necessary for replicating this information. The genetic material of the influenza virus is a single-stranded negative-sense RNA (ss-RNA). This ss-RNA binds with nucleoproteins (NP), coiling into ribonucleoprotein (RNP) complexes, existing in a highly dense form. Apart from RNPs, there is also an RNA polymerase responsible for RNA transcription. Both Type A and Type B influenza viruses' RNA is composed of 8 segments, while Type C has one less.
Matrix Protein The matrix protein forms the virus's outer shell. In reality, besides matrix protein (M1), there is also membrane protein (M2). The matrix protein binds tightly with the outermost envelope of the virus, playing a role in protecting the virus core and maintaining its spatial structure.
Envelope The envelope is a phospholipid bilayer membrane outside the matrix protein. Mature influenza viruses bud from host cells, wrapping the host's cell membrane around themselves and then detaching to infect the next target. Within the envelope, besides phospholipid molecules, there are two crucial glycoproteins: hemagglutinin and neuraminidase.
Hemagglutinin (HA) It's rod-shaped and can bind with receptors on the surface of red blood cells from humans, birds, pigs, and other animals, causing clotting, hence the name "hemagglutinin".
Neuraminidase (NA) This is a mushroom-shaped tetramer glycoprotein with the activity of hydrolyzing sialic acid. After mature influenza viruses bud from host cells, neuraminidase plays a crucial role in the release of viruses from host cells. This enzyme is also a target for influenza treatment drugs, with Oseltamivir being one of the most famous anti-influenza drugs targeting it.
流行性感冒病毒简称流感病毒。它分为甲(A)、乙(B)、丙(C)三型,近年来才发现的流感病毒将归为丁(D)型。流感病毒可引起人、禽、猪、马、蝙蝠等多种动物感染和发病,是人流感、禽流感、猪流感、马流感等人与动物疫病的病原。
流行性感冒病毒(influenza virus),是正粘病毒科(Orthomyxoviridae)的代表种,简称流感病毒,包括人流感病毒和动物流感病毒,人流感病毒分为甲(A)、乙(B)、丙(C)三型,是流行性感冒(流感)的病原体。其中甲型流感病毒抗原性易发生变异,多次引起世界性大流行。例如1918~1919年的大流行中,全世界至少有2000万~4000万人死于流感;乙型流感病毒对人类致病性也比较强,但是人们还没有发现乙型流感病毒引起过世界性大流行;丙型流感病毒只引起人类不明显的或轻微的上呼吸道感染,很少造成流行。甲型流感病毒于1933年分离成功,乙型流感病毒于1940年获得,丙型流感病毒直到1949年才成功分离。
流感分类
根据流感病毒感染的对象,可以将病毒分为人类流感病毒、猪流感病毒、马流感病毒以及禽流感病毒等类群,其中人类流感病毒根据其核蛋白的抗原性可以分为三类:
甲型流感病毒(Influenza A virus),又称A型流感病毒
乙型流感病毒(Influenza B virus),又称B型流感病毒
丙型流感病毒(Influenza C virus),又称C型流感病毒
感染鸟类、猪等其他动物的流感病毒,其核蛋白的抗原性与人甲型流感病毒相同,但是由于甲型、乙型和丙型流感病毒的分类只是针对人流感病毒的,因此通常不将禽流感病毒等非人类宿主的流感病毒称作甲型流感病毒。
在核蛋白抗原性的基础上,流感病毒还根据血凝素HA和神经氨酸酶NA的抗原性分为不同的亚型。
形态结构
流感病毒呈球形,新分离的毒株则多呈丝状,其直径在80至120纳米之间,丝状流感病毒的长度可达4000纳米。
流感病毒结构自外而内可分为包膜、基质蛋白以及核心三部分。
核心
病毒的核心包含了存贮病毒信息的遗传物质以及复制这些信息必须的酶。流感病毒的遗传物质是单股负链RNA,简写为ss-RNA,ss-RNA与核蛋白 (NP)相结合,缠绕成核糖核蛋白体(RNP),以密度极高的形式存在。除了核糖核蛋白体,还有负责RNA转录的RNA多聚酶。
甲型和乙型流感病毒的RNA由8个节段组成,丙型流感病毒则比它们少一个节段,第1、2、3个节段编码的是RNA多聚集酶,第4个节段负责编码血凝素;第5个节段负责编码核蛋白,第6个节段编码的是神经氨酸酶;第7个节段编码基质蛋白,第8个节段编码的是一种能起到拼接RNA功能的非结构蛋白,这种蛋白的其他功能尚不得而知。
丙型流感病毒缺少的是第六个节段,其第四节段编码的血凝素可以同时行使神经氨酸酶的功能。
基质蛋白
基质蛋白构成了病毒的外壳骨架,实际上骨架中除了基质蛋白 (M1)之外还有膜蛋白 (M2)。M2蛋白具有离子(主要是Na+)通道和调节膜内PH值的作用,但数量很少。基质蛋白与病毒最外层的包膜紧密结合起到保护病毒核心和维系病毒空间结构的作用。
当流感病毒在宿主细胞内完成其繁殖之后,基质蛋白是分布在宿主细胞细胞膜内壁上的,成型的病毒核衣壳能够识别宿主细胞膜上含有基质蛋白的部位,与之结合形成病毒结构,并以出芽的形式突出释放成熟病毒。
包膜
包膜是包裹在基质蛋白之外的一层磷脂双分子层膜,这层膜来源于宿主的细胞膜,成熟的流感病毒从宿主细胞出芽,将宿主的细胞膜包裹在自己身上之后脱离细胞,去感染下一个目标。
包膜中除了磷脂分子之外,还有两种非常重要的糖蛋白:血凝素和神经氨酸酶。这两类蛋白突出病毒体外,长度约为10至40纳米,被称作刺突。一般一个流感病毒表面会分布有500个血凝素刺突和100个神经氨酸酶刺突。在甲型流感病毒中血凝素和神经氨酸酶的抗原性会发生变化,这是区分病毒毒株亚型的依据。
血凝素(HA)
呈柱状,能与人、鸟、猪豚鼠等动物红细胞表面的受体相结合引起凝血,故而被称作血凝素。血凝素蛋白水解后分为轻链和重链两部分,后者可以与宿主细胞膜上的唾液酸受体相结合,前者则可以协助病毒包膜与宿主细胞膜相互融合。血凝素在病毒导入宿主细胞的过程中扮演了重要角色。血凝素具有免疫原性,抗血凝素抗体可以中和流感病毒。
神经氨酸酶(NA)
是一个呈蘑菇状的四聚体糖蛋白,具有水解唾液酸的活性,当成熟的流感病毒经出芽的方式脱离宿主细胞之后,病毒表面的血凝素会经由唾液酸受体与宿主细胞膜保持联系,需要由神经氨酸酶将唾液酸水解,切断病毒与宿主细胞的最后联系,使病毒能顺利从宿主细胞中释放,继而感染下一个宿主细胞。因此神经氨酸酶也成为流感治疗药物的一个作用靶点,针对此酶设计的奥司他韦是最著名的抗流感药物之一。





